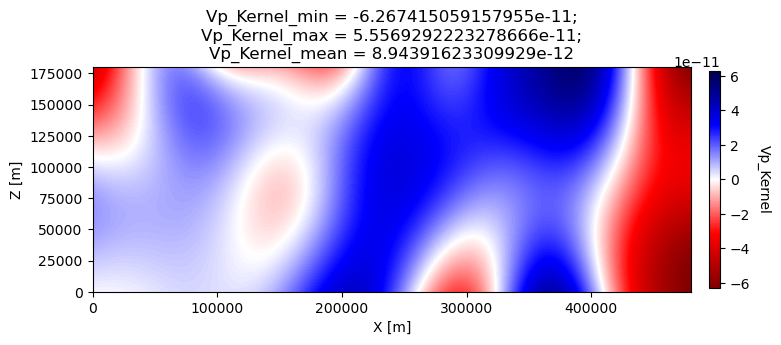
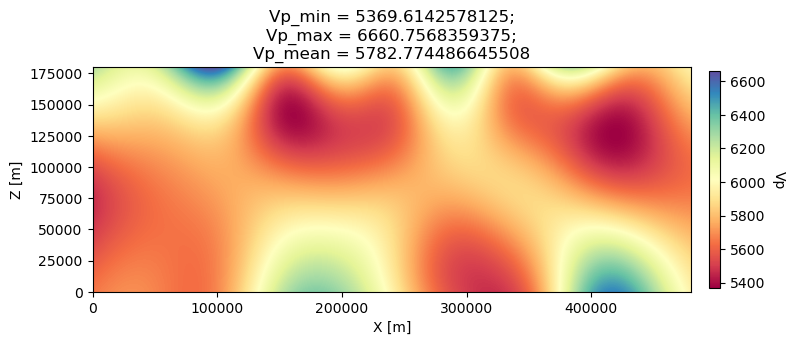
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
| #-----------------------------------------------------------
#
#
# title of job
title = Tape-Liu-Tromp (GJI 2007)
# forward or adjoint simulation
# 1 = forward, 2 = adjoint, 3 = both simultaneously
# note: 2 is purposely UNUSED (for compatibility with the numbering of our 3D codes)
SIMULATION_TYPE = 1
# 0 = regular wave propagation simulation, 1/2/3 = noise simulation
NOISE_TOMOGRAPHY = 0
# save the last frame, needed for adjoint simulation
SAVE_FORWARD = .false.
# parameters concerning partitioning
NPROC = 4 # number of processes
# time step parameters
# total number of time steps
NSTEP = 14000
# duration of a time step (see section "How to choose the time step" of the manual for how to do this)
DT = 2.5d-2
# time stepping
# 1 = Newmark (2nd order), 2 = LDDRK4-6 (4th-order 6-stage low storage Runge-Kutta), 3 = classical RK4 4th-order 4-stage Runge-Kutta
time_stepping_scheme = 1
# set the type of calculation (P-SV or SH/membrane waves)
P_SV = .true.
# axisymmetric (2.5D) or Cartesian planar (2D) simulation
AXISYM = .false.
#-----------------------------------------------------------
#
#
# Partitioning algorithm for decompose_mesh
PARTITIONING_TYPE = 3 # SCOTCH = 3, ascending order (very bad idea) = 1
# number of control nodes per element (4 or 9)
NGNOD = 4
# creates/reads a binary database that allows to skip all time consuming setup steps in initialization
# 0 = does not read/create database
# 1 = creates database
# 2 = reads database
setup_with_binary_database = 1
# available models
# default - define model using nbmodels below
# ascii - read model from ascii database file
# binary - read model from binary databse file
# binary_voigt - read Voigt model from binary database file
# external - define model using define_external_model subroutine
# gll - read GLL model from binary database file
# legacy - read model from model_velocity.dat_input
MODEL = gll
# Output the model with the requested type, does not save if turn to default or .false.
# (available output formats: ascii,binary,gll,legacy)
SAVE_MODEL = binary
#-----------------------------------------------------------
#
#
# attenuation parameters
ATTENUATION_VISCOELASTIC = .false. # turn attenuation (viscoelasticity) on or off for non-poroelastic solid parts of the model
ATTENUATION_VISCOACOUSTIC = .false. # turn attenuation (viscoacousticity) on or off for non-poroelastic fluid parts of the model
# for viscoelastic or viscoacoustic attenuation
N_SLS = 3 # number of standard linear solids for attenuation (3 is usually the minimum)
ATTENUATION_f0_REFERENCE = 5.196152422706633 # in case of attenuation, reference frequency in Hz at which the velocity values in the velocity model are given (unused otherwise); relevant only if source is a Dirac or a Heaviside, otherwise it is automatically set to f0 the dominant frequency of the source in the DATA/SOURCE file
READ_VELOCITIES_AT_f0 = .false. # read seismic velocities at ATTENUATION_f0_REFERENCE instead of at infinite frequency (see user manual for more information)
USE_SOLVOPT = .false. # use more precise but much more expensive way of determining the Q factor relaxation times, as in https://doi.org/10.1093/gji/ggw024
# for poroelastic attenuation
ATTENUATION_PORO_FLUID_PART = .false. # turn viscous attenuation on or off for the fluid part of poroelastic parts of the model
Q0_poroelastic = 1 # quality factor for viscous attenuation (ignore it if you are not using a poroelastic material)
freq0_poroelastic = 10 # frequency for viscous attenuation (ignore it if you are not using a poroelastic material)
# to undo attenuation and/or PMLs for sensitivity kernel calculations or forward runs with SAVE_FORWARD
# use the flag below. It performs undoing of attenuation and/or of PMLs in an exact way for sensitivity kernel calculations
# but requires disk space for temporary storage, and uses a significant amount of memory used as buffers for temporary storage.
# When that option is on the second parameter indicates how often the code dumps restart files to disk (if in doubt, use something between 100 and 1000).
UNDO_ATTENUATION_AND_OR_PML = .false.
NT_DUMP_ATTENUATION = 500
# Instead of reconstructing the forward wavefield, this option reads it from the disk using asynchronous I/O.
# Outperforms conventional mode using a value of NTSTEP_BETWEEN_COMPUTE_KERNELS high enough.
NO_BACKWARD_RECONSTRUCTION = .false.
#-----------------------------------------------------------
#
#
# source parameters
NSOURCES = 1 # number of sources (source information is then read from the DATA/SOURCE file)
force_normal_to_surface = .false. # angleforce normal to surface (external mesh and curve file needed)
# use an existing initial wave field as source or start from zero (medium initially at rest)
initialfield = .false.
add_Bielak_conditions_bottom = .false. # add Bielak conditions or not if initial plane wave
add_Bielak_conditions_right = .false.
add_Bielak_conditions_top = .false.
add_Bielak_conditions_left = .false.
# acoustic forcing
ACOUSTIC_FORCING = .false. # acoustic forcing of an acoustic medium with a rigid interface
# noise simulations - type of noise source time function:
# 0=external (S_squared), 1=Ricker(second derivative), 2=Ricker(first derivative), 3=Gaussian, 4=Figure 2a of Tromp et al. 2010
# (default value 4 is chosen to reproduce the time function from Fig 2a of "Tromp et al., 2010, Noise Cross-Correlation Sensitivity Kernels")
noise_source_time_function_type = 4
# moving sources
# Set write_moving_sources_database to .true. if the generation of moving source databases takes
# a long time. Then the simulation is done in two steps: first you run the code and it writes the databases to file
# (in DATA folder by default). Then you rerun the code and it will read the databases in there directly possibly
# saving a lot of time.
# This is only useful for GPU version (for now)
write_moving_sources_database = .false.
#-----------------------------------------------------------
#
#
# receiver set parameters for recording stations (i.e. recording points)
# seismotype : record 1=displ 2=veloc 3=accel 4=pressure 5=curl of displ 6=the fluid potential
seismotype = 1 # several values can be chosen. For example : 1,2,4
# interval in time steps for writing of seismograms
# every how many time steps we save the seismograms
# (costly, do not use a very small value; if you use a very large value that is larger than the total number
# of time steps of the run, the seismograms will automatically be saved once at the end of the run anyway)
NTSTEP_BETWEEN_OUTPUT_SEISMOS = 100
# set to n to reduce the sampling rate of output seismograms by a factor of n
# defaults to 1, which means no down-sampling
NTSTEP_BETWEEN_OUTPUT_SAMPLE = 1
# so far, this option can only be used if all the receivers are in acoustic elements
USE_TRICK_FOR_BETTER_PRESSURE = .false.
# use this t0 as earliest starting time rather than the automatically calculated one
USER_T0 = 90.0d0
# seismogram formats
save_ASCII_seismograms = .true. # save seismograms in ASCII format or not
save_binary_seismograms_single = .false. # save seismograms in single precision binary format or not (can be used jointly with ASCII above to save both)
save_binary_seismograms_double = .false. # save seismograms in double precision binary format or not (can be used jointly with both flags above to save all)
SU_FORMAT = .false. # output single precision binary seismograms in Seismic Unix format (adjoint traces will be read in the same format)
# use an existing STATION file found in ./DATA or create a new one from the receiver positions below in this Par_file
use_existing_STATIONS = .false.
# number of receiver sets (i.e. number of receiver lines to create below)
nreceiversets = 1
# orientation
anglerec = 0.d0 # angle to rotate components at receivers
rec_normal_to_surface = .false. # base anglerec normal to surface (external mesh and curve file needed)
# first receiver set (repeat these 6 lines and adjust nreceiversets accordingly)
nrec = 40 # number of receivers
xdeb = 10000. # first receiver x in meters
zdeb = 180000. # first receiver z in meters
xfin = 470000. # last receiver x in meters (ignored if only one receiver)
zfin = 180000. # last receiver z in meters (ignored if only one receiver)
record_at_surface_same_vertical = .true. # receivers inside the medium or at the surface
#-----------------------------------------------------------
#
#
# save sensitivity kernels in ASCII format (much bigger files, but compatible with current GMT scripts) or in binary format
save_ASCII_kernels = .false.
# since the accuracy of kernel integration may not need to respect the CFL, this option permits to save computing time, and memory with UNDO_ATTENUATION_AND_OR_PML mode
NTSTEP_BETWEEN_COMPUTE_KERNELS = 1
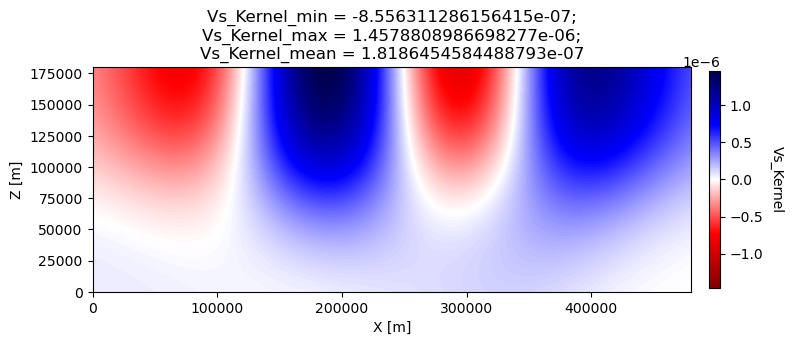
# outputs approximate Hessian for preconditioning
APPROXIMATE_HESS_KL = .false.
#-----------------------------------------------------------
#
#
# Perfectly Matched Layer (PML) boundaries
# absorbing boundary active or not
PML_BOUNDARY_CONDITIONS = .false.
NELEM_PML_THICKNESS = 3
ROTATE_PML_ACTIVATE = .false.
ROTATE_PML_ANGLE = 30.
# change the four parameters below only if you know what you are doing; they change the damping profiles inside the PMLs
K_MIN_PML = 1.0d0 # from Gedney page 8.11
K_MAX_PML = 1.0d0
damping_change_factor_acoustic = 0.5d0
damping_change_factor_elastic = 1.0d0
# set the parameter below to .false. unless you know what you are doing; this implements automatic adjustment of the PML parameters for elongated models.
# The goal is to improve the absorbing efficiency of PML for waves with large incidence angles, but this can lead to artefacts.
# In particular, this option is efficient only when the number of sources NSOURCES is equal to one.
PML_PARAMETER_ADJUSTMENT = .false.
# Stacey ABC
STACEY_ABSORBING_CONDITIONS = .true.
# periodic boundaries
ADD_PERIODIC_CONDITIONS = .false.
PERIODIC_HORIZ_DIST = 0.3597d0
#-----------------------------------------------------------
#
#
# number of model materials
nbmodels = 2
# available material types (see user manual for more information)
# acoustic: model_number 1 rho Vp 0 0 0 QKappa 9999 0 0 0 0 0 0 (for QKappa use 9999 to ignore it)
# elastic: model_number 1 rho Vp Vs 0 0 QKappa Qmu 0 0 0 0 0 0 (for QKappa and Qmu use 9999 to ignore them)
# anisotropic: model_number 2 rho c11 c13 c15 c33 c35 c55 c12 c23 c25 0 QKappa Qmu
# anisotropic in AXISYM: model_number 2 rho c11 c13 c15 c33 c35 c55 c12 c23 c25 c22 QKappa Qmu
# poroelastic: model_number 3 rhos rhof phi c kxx kxz kzz Ks Kf Kfr etaf mufr Qmu
# tomo: model_number -1 0 0 A 0 0 0 0 0 0 0 0 0 0
#
# the Vp and Vs values that are read here are the UNRELAXED ones i.e. the values at infinite frequency
# unless the READ_VELOCITIES_AT_f0 parameter above is set to true, in which case they are the values at frequency f0.
#
# To convert one to the other see doc/Qkappa_Qmu_versus_Qp_Qs_relationship_in_2D_plane_strain.pdf and
# utils/attenuation/conversion_from_Qkappa_Qmu_to_Qp_Qs_from_Dahlen_Tromp_959_960.f90.
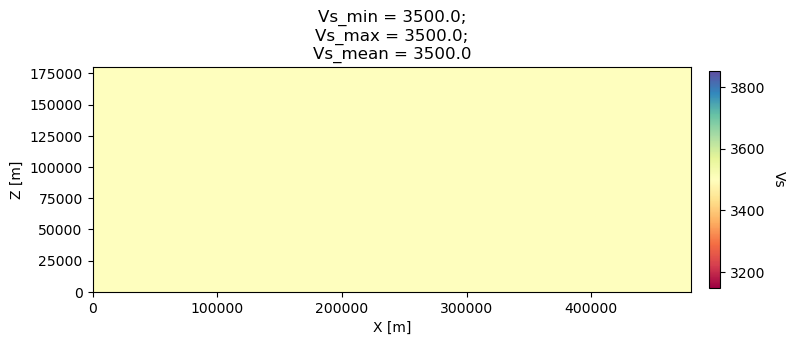
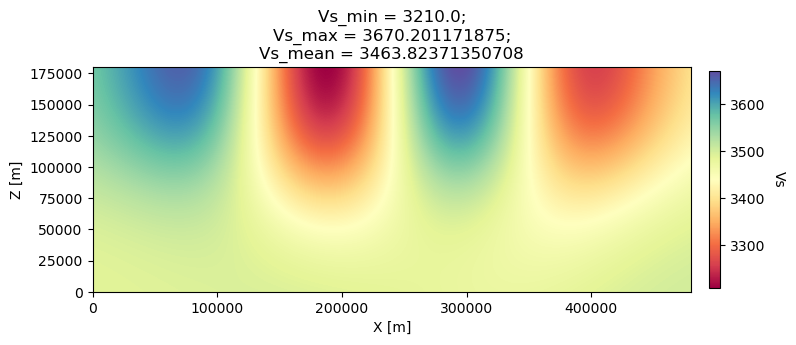
1 1 2600.d0 5800.d0 3500.0d0 0 0 10.d0 10.d0 0 0 0 0 0 0
2 1 2600.d0 5800.d0 3500.0d0 0 0 10.d0 10.d0 0 0 0 0 0 0
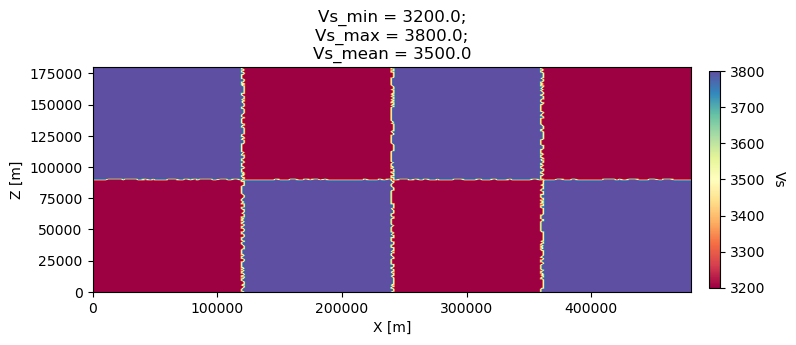
#1 1 2600.d0 5600.d0 3200.0d0 0 0 10.d0 10.d0 0 0 0 0 0 0
#2 1 2600.d0 6000.d0 3800.0d0 0 0 10.d0 10.d0 0 0 0 0 0 0
# external tomography file
TOMOGRAPHY_FILE = ./DATA/tomo_file.xyz
# use an external mesh created by an external meshing tool or use the internal mesher
read_external_mesh = .false.
#-----------------------------------------------------------
#
#
#-----------------------------------------------------------
# data concerning mesh, when generated using third-party app (more info in README)
# (see also absorbing_conditions above)
mesh_file = ./DATA/ice_water_rock_1D/ice_water_rock_1D_elements # file containing the mesh
nodes_coords_file = ./DATA/ice_water_rock_1D/ice_water_rock_1D_nodes # file containing the nodes coordinates
materials_file = ./DATA/ice_water_rock_1D/ice_water_rock_1D_material # file containing the material number for each element
free_surface_file = ./DATA/ice_water_rock_1D/ice_water_rock_1D_surface_free # file containing the free surface
axial_elements_file = ./DATA/axial_elements_file # file containing the axial elements if AXISYM is true
absorbing_surface_file = ./DATA/ice_water_rock_1D/ice_water_rock_1D_surface_absorb # file containing the absorbing surface
acoustic_forcing_surface_file = ./DATA/MSH/Surf_acforcing_Bottom_enforcing_mesh # file containing the acoustic forcing surface
absorbing_cpml_file = ./DATA/absorbing_cpml_file # file containing the CPML element numbers
tangential_detection_curve_file = ./DATA/courbe_eros_nodes # file containing the curve delimiting the velocity model
#-----------------------------------------------------------
#
#
# file containing interfaces for internal mesh
interfacesfile = interfaces_Tape2007.dat
# geometry of the model (origin lower-left corner = 0,0) and mesh description
xmin = 0.d0 # abscissa of left side of the model
xmax = 480000.d0 # abscissa of right side of the model
nx = 40 # number of elements along X
# absorbing boundary parameters (see absorbing_conditions above)
absorbbottom = .false.
absorbright = .true.
absorbtop = .true.
absorbleft = .true.
# define the different regions of the model in the (nx,nz) spectral-element mesh
nbregions = 8 # then set below the different regions and model number for each region
# format of each line: nxmin nxmax nzmin nzmax material_number
1 10 1 20 1
11 20 1 20 2
21 30 1 20 1
31 40 1 20 2
1 10 21 40 2
11 20 21 40 1
21 30 21 40 2
31 40 21 40 1
#-----------------------------------------------------------
#
#
# interval at which we output time step info and max of norm of displacement
# (every how many time steps we display information about the simulation. costly, do not use a very small value)
NTSTEP_BETWEEN_OUTPUT_INFO = 400
# meshing output
output_grid_Gnuplot = .false. # generate a GNUPLOT file containing the grid, and a script to plot it
output_grid_ASCII = .false. # dump the grid in an ASCII text file consisting of a set of X,Y,Z points or not
# to plot total energy curves, for instance to monitor how CPML absorbing layers behave;
# should be turned OFF in most cases because a bit expensive
OUTPUT_ENERGY = .false.
# every how many time steps we compute energy (which is a bit expensive to compute)
NTSTEP_BETWEEN_OUTPUT_ENERGY = 10
# Compute the field int_0^t v^2 dt for a set of GLL points and write it to file. Use
# the script utils/visualisation/plotIntegratedEnergyFile.py to watch. It is refreshed at the same time than the seismograms
COMPUTE_INTEGRATED_ENERGY_FIELD = .false.
#-----------------------------------------------------------
#
#
# every how many time steps we draw JPEG or PostScript pictures of the simulation
# and/or we dump results of the simulation as ASCII or binary files (costly, do not use a very small value)
NTSTEP_BETWEEN_OUTPUT_IMAGES = 200
# minimum amplitude kept in % for the JPEG and PostScript snapshots; amplitudes below that are muted
cutsnaps = 1.
#
output_color_image = .true. # output JPEG color image of the results every NTSTEP_BETWEEN_OUTPUT_IMAGES time steps or not
imagetype_JPEG = 3 # display 1=displ_Ux 2=displ_Uz 3=displ_norm 4=veloc_Vx 5=veloc_Vz 6=veloc_norm 7=accel_Ax 8=accel_Az 9=accel_norm 10=pressure
factor_subsample_image = 1.0d0 # (double precision) factor to subsample or oversample (if set to e.g. 0.5) color images output by the code (useful for very large models, or to get nicer looking denser pictures)
USE_CONSTANT_MAX_AMPLITUDE = .false. # by default the code normalizes each image independently to its maximum; use this option to use the global maximum below instead
CONSTANT_MAX_AMPLITUDE_TO_USE = 1.17d4 # constant maximum amplitude to use for all color images if the above USE_CONSTANT_MAX_AMPLITUDE option is true
POWER_DISPLAY_COLOR = 0.30d0 # non linear display to enhance small amplitudes in JPEG color images
DRAW_SOURCES_AND_RECEIVERS = .true. # display sources as orange crosses and receivers as green squares in JPEG images or not
DRAW_WATER_IN_BLUE = .true. # display acoustic layers as constant blue in JPEG images, because they likely correspond to water in the case of ocean acoustics or in the case of offshore oil industry experiments (if off, display them as greyscale, as for elastic or poroelastic elements, for instance for acoustic-only oil industry models of solid media)
USE_SNAPSHOT_NUMBER_IN_FILENAME = .false. # use snapshot number in the file name of JPEG color snapshots instead of the time step (for instance to create movies in an easier way later)
#
output_postscript_snapshot = .false. # output Postscript snapshot of the results every NTSTEP_BETWEEN_OUTPUT_IMAGES time steps or not
imagetype_postscript = 1 # display 1=displ vector 2=veloc vector 3=accel vector; small arrows are displayed for the vectors
meshvect = .false. # display mesh on PostScript plots or not
modelvect = .false. # display velocity model on PostScript plots or not
boundvect = .true. # display boundary conditions on PostScript plots or not
interpol = .true. # interpolation of the PostScript display on a regular grid inside each spectral element, or use the non-evenly spaced GLL points
pointsdisp = 6 # number of points in each direction for interpolation of PostScript snapshots (set to 1 for lower-left corner only)
subsamp_postscript = 1 # subsampling of background velocity model in PostScript snapshots
sizemax_arrows = 1.d0 # maximum size of arrows on PostScript plots in centimeters
US_LETTER = .false. # use US letter or European A4 paper for PostScript plots
#
output_wavefield_dumps = .false. # output wave field to a text file (creates very big files)
imagetype_wavefield_dumps = 1 # display 1=displ vector 2=veloc vector 3=accel vector 4=pressure
use_binary_for_wavefield_dumps = .false. # use ASCII or single-precision binary format for the wave field dumps
#-----------------------------------------------------------
# Ability to run several calculations (several earthquakes)
# in an embarrassingly-parallel fashion from within the same run;
# this can be useful when using a very large supercomputer to compute
# many earthquakes in a catalog, in which case it can be better from
# a batch job submission point of view to start fewer and much larger jobs,
# each of them computing several earthquakes in parallel.
# To turn that option on, set parameter NUMBER_OF_SIMULTANEOUS_RUNS to a value greater than 1.
# To implement that, we create NUMBER_OF_SIMULTANEOUS_RUNS MPI sub-communicators,
# each of them being labeled "my_local_mpi_comm_world", and we use them
# in all the routines in "src/shared/parallel.f90", except in MPI_ABORT() because in that case
# we need to kill the entire run.
# When that option is on, of course the number of processor cores used to start
# the code in the batch system must be a multiple of NUMBER_OF_SIMULTANEOUS_RUNS,
# all the individual runs must use the same number of processor cores,
# which as usual is NPROC in the Par_file,
# and thus the total number of processor cores to request from the batch system
# should be NUMBER_OF_SIMULTANEOUS_RUNS * NPROC.
# All the runs to perform must be placed in directories called run0001, run0002, run0003 and so on
# (with exactly four digits).
#
#
#
# each running on 100 cores (see e.g. http://www.schedmd.com/slurmdocs/job_array.html )
#
# cd into one of 10 subdirectories (called e.g. run0001, run0002,... run0010) depending on the sub-communicator
# your MPI rank belongs to, and run normally on 100 cores using that sub-communicator.
#
#
NUMBER_OF_SIMULTANEOUS_RUNS = 1
# if we perform simultaneous runs in parallel, if only the source and receivers vary between these runs
# but not the mesh nor the model (velocity and density) then we can also read the mesh and model files
# from a single run in the beginning and broadcast them to all the others; for a large number of simultaneous
# runs for instance when solving inverse problems iteratively this can DRASTICALLY reduce I/Os to disk in the solver
# (by a factor equal to NUMBER_OF_SIMULTANEOUS_RUNS), and reducing I/Os is crucial in the case of huge runs.
# Thus, always set this option to .true. if the mesh and the model are the same for all simultaneous runs.
# In that case there is no need to duplicate the mesh and model file database (the content of the DATABASES_MPI
# directories) in each of the run0001, run0002,... directories, it is sufficient to have one in run0001
# and the code will broadcast it to the others)
BROADCAST_SAME_MESH_AND_MODEL = .true.
#-----------------------------------------------------------
# set to true to use GPUs
GPU_MODE = .false.
|